


There have been numerous benchmarks posted around the web and in papers outlining how CRAM stacks up against other file formats. While I fully encourage such comparisons, it should be noted that CRAM is not a single format. Even within a single implementation there can be many ways to produce a valid CRAM file, and between implementations there may be substantial differences in both file size and speed. This can lead to accidents, creating misleading statistics that do not fully show the broad spectrum of CRAM performance. One such example is Spiral Genetics SpEC format (and the primary reason for posting this, given several people have prompted me to look at their site, http://www.spiralgenetics.com/spec/results/).
To quote their comparison: “To compare methods, we compress a standard 50x NA12878 data set. The original data, in BAM format, is 114GB. CRAM was run in lossless mode. Both analyses were run on a machine with 32 GB RAM and 16 cores”. Similarly I chose a 16 core machine, albeit an older perhaps slower one; a 16 core Xeon E5-2660 running at 2.2GHz. I gave 16 cores available to all of the tools I benchmarked, but note that not all tools fully support multi-threading. This seems like an unfair comparison, but it is reproducing the same conditions in the SpEC benchmarks. For fairness I also show the total CPU time, which gives an indication of single-threaded performance and may hint at how programs could compare if all were multi-threaded. The input data set is Illumina Platinum Genomes NA12878, from EBI: ftp://ftp.sra.ebi.ac.uk/vol1/ERA172/ERA172924/bam/NA12878_S1.bam.
Firstly, file size. The following reflects combinations of program (Samtools, Scramble, Cramtools, Deez, and (assumed name) spec2bam) along with file formats used within those programs (BAM, CRAM, DZ and SPEC). CRAM++ is an experimental branch of Staden io_lib that uses some custom codecs taken out of fqzcomp. It is not intended for production use, but simply to act as a data point to show how the file size / time tradeoff works if we are willing to throw more CPU at the problem. There are two Scramble/CRAM entries here. The first is the default mode while the second is with parameters “-7 -j -Z”. These indicate a higher level of compression plus the ability to use bzip and/or lzma compression if required. This is permitted within the CRAM specification (unlike the CRAM++ entry), but is not the default as the size/time trade off may not be worth it for many people. Samtools/htslib also supports CRAM, but for brevity I did not benchmark it here. It will be almost identical in performance to Scramble/CRAM once Samtools-1.3 is released.

Note how this differs substantially to the published SpEC results. The above is with CRAM version 3.0, while the SpEC results were produced using Cramtools-2.1.jar. SpEC is smaller than CRAM v3 by 1-2%, although larger than the experimental codecs. Java Cramtools.jar is still undergoing optimisation of CRAM v3 codecs, so produces files larger than Scramble/Samtools. The version 3.0 CRAM files produced by scramble, samtools and cramtools are all interchangeable, so this is a practical demonstration of a way to mislead yourself. Don't just check the file format, but check the program/format combination – or better program/format/version combination as almost certainly cramtools will be producing smaller files soon.
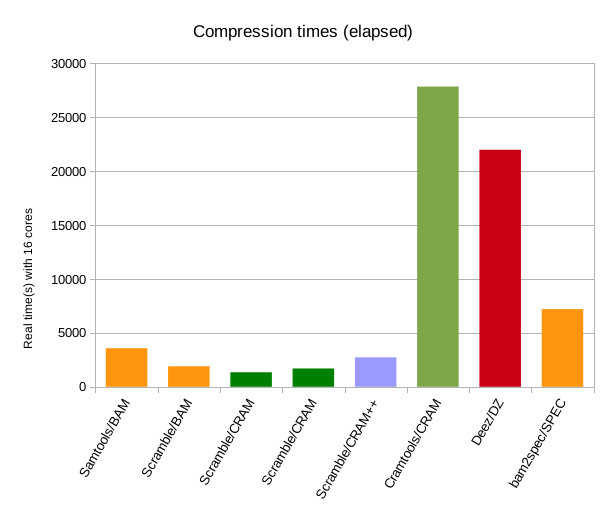
The above left graph is comparable to the SpEC results, showing Cramools/CRAM performance against SpEC performance. Note that the SPEC timings here are taken from their web page (“around 2 hours”) and are likely favourably compared given the relative slowness of this machine – they had Cramtools taking 7 hours while it is closer to 8 on this system. Also note the difference between Samtools/BAM and Scramble/BAM. This isn't due to any magic version of zlib, but simply that Samtools does not currently support multi-threaded decoding, so the process becomes bottlenecked on decoding the input BAM. Fixing this is something that the Samtools team have on their wish-list.
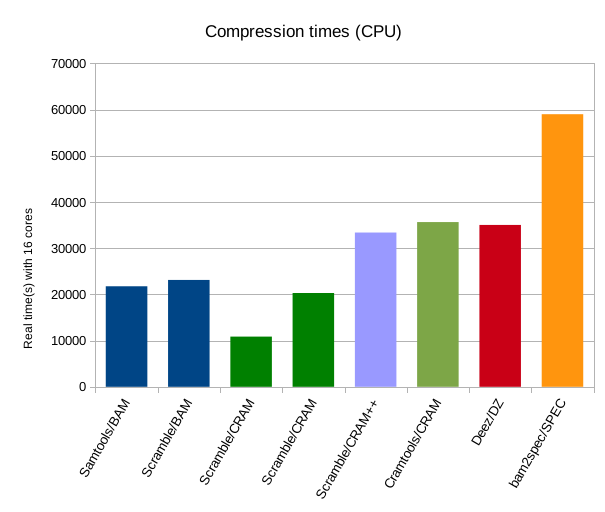
The above right graph shows total CPU time (user + system). Much clearer hear is the difference in CPU (or single-threaded real time) between basic CRAM and CRAM with bzip / lzma codecs. Also clear is how much more time the fqzcomp codecs would consume, both elapsed and CPU seconds, however it is still comparable to Deez and under half the CPU cost of SpEC.
Comparing the elapsed vs CPU times it is also evident how unfair it is to only show elapsed timings without note to which tools support multi-threading. The lack of significant multi-threading is very obvious in Cramtools and Deez, but we can get an idea of the expected performance from these tools if multi-threaded was added. (I fully expect this to appear in Cramtools in a future release.)


Decode timings here are the time taken to read the BAM/CRAM/DZ/SPEC file and emit an uncompressed BAM (Samtools, Scramble, and Spec2Bam) or an uncompressed SAM (Deez and Cramtools) file. Clearly it is slower to output in uncompressed SAM than BAM, so this needs to be taken into consideration. Ideally I would have used uncompressed BAM throughout, but this was not an option.
Decompression times are more extreme. As noted already, Samtools currently lacks multi-threaded decoding, which is why it is comparable CPU time to Scramble but more elasped time. However both tools decode BAM faster than any other format, including CRAM. It is evident from this test though that the Scramble/Samtools C implementation of CRAM would beat Spec2Bam in speed even if we compared a single thread against 16.
Encode |
Decode |
||||||
|---|---|---|---|---|---|---|---|
Program |
Format |
Arguments |
File Size |
Real time(s) |
CPU time(s) |
Real time(s) |
CPU time(s) |
Samtools |
BAM |
-@16 -b |
121709642739 |
3583 |
21746 |
2172 |
4936 |
Scramble |
BAM |
-t16 -O bam |
121694779760 |
1901 |
23123 |
474 |
5294 |
Scramble |
CRAM |
-t16 -O cram -r hg19.fa |
66613992539 |
1349 |
10857 |
752 |
6485 |
Scramble |
CRAM |
-t16 -O cram -r hg19.fa -7 -jZ |
65196595346 |
1698 |
20300 |
744 |
8543 |
Scramble |
CRAM++ |
-t16 -O cram -r hg19.fa |
59728988392 |
2734 |
33391 |
1301 |
21316 |
Cramtools |
CRAM |
--capture-all-tags -Q -n -r hg19.fa |
72366049509 |
27857 |
35658 |
29026 |
30400 |
Deez |
DZ |
-t16 -r hg19.fa |
68923106569 |
21991 |
35042 |
12030 |
19347 |
Spec? |
SPEC |
--ref hg19.fa --threads 16 |
64775870649 |
7200 |
59000 |
10152 |
83362 |
I have shown the performance of CRAM, both file size and elapsed / CPU times, varies significantly between implementations. Please do remember this when attempting to benchmark against it.
James Bonfield, Wellcome Trust Sanger Institute.

